49 Predicting with Regression
49.1 Key Ideas
- Fit a simple regression model
- Plug in new covariates and multiply them by the coefficients
- Useful when the linear model is (nearly) correct
Pros: * Easy to implement * easy to interpret
Cons: * Often poor performance on non-linear settings
49.2 Example: Geyser Eruptions
library(caret); data(faithful); set.seed(1234)
inTrain <- createDataPartition(y = faithful$waiting,
p = 0.5, list = FALSE)
trainFaith <- faithful[inTrain, ];
testFaith <- faithful[-inTrain, ]
head(trainFaith)## eruptions waiting
## 6 2.883 55
## 7 4.700 88
## 9 1.950 51
## 11 1.833 54
## 12 3.917 84
## 13 4.200 78# Plot the waiting vs eruption data
ggplot(data = trainFaith, aes(x = waiting, y = eruptions, col = eruptions)) +
geom_point() +
geom_smooth( method = "lm")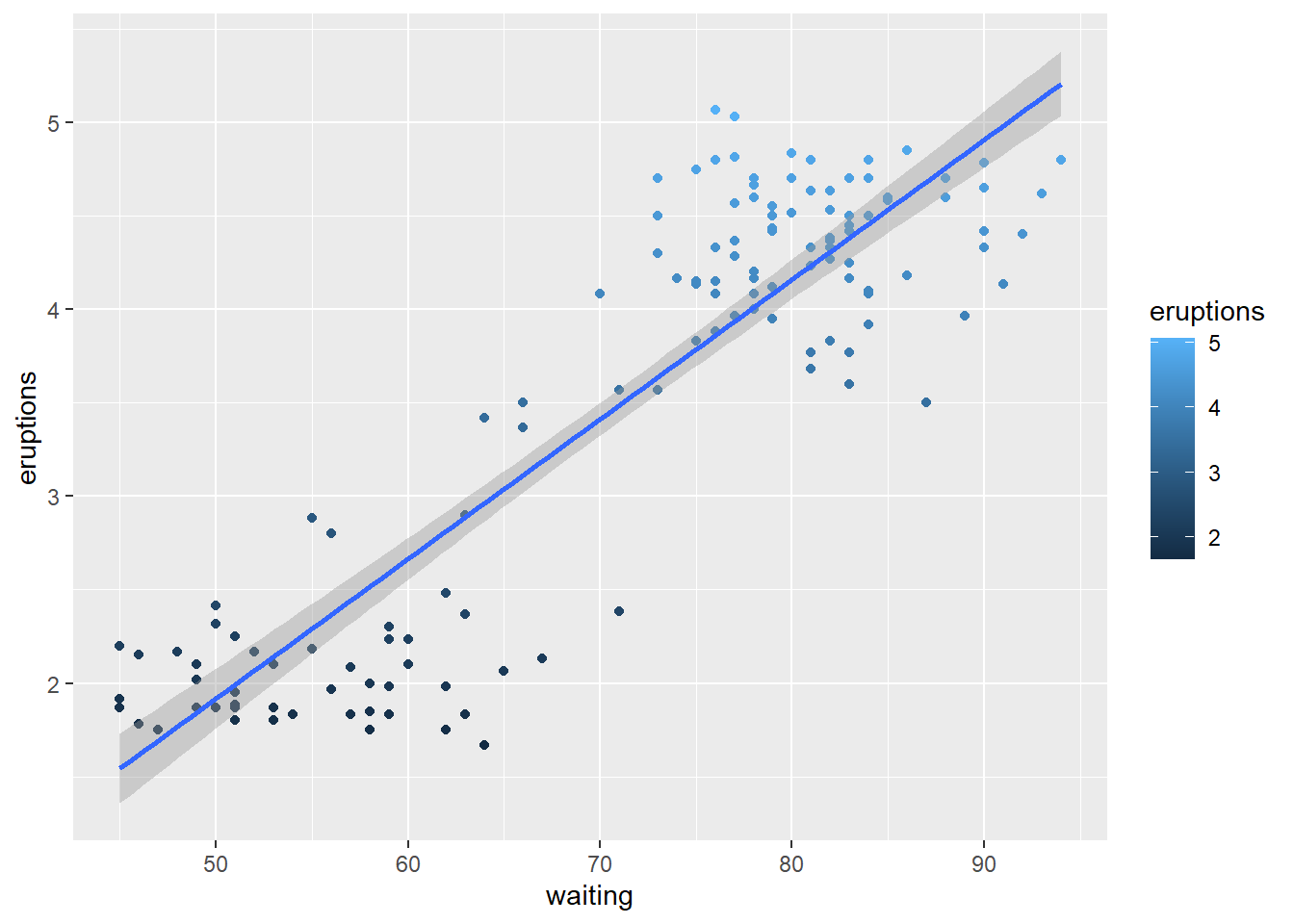
# Lets make an example linear model predicting eruptions using waiting time
lm1 <- lm(eruptions ~ waiting, data = trainFaith)
summary(lm1)##
## Call:
## lm(formula = eruptions ~ waiting, data = trainFaith)
##
## Residuals:
## Min 1Q Median 3Q Max
## -1.29776 -0.37733 0.03967 0.35578 1.20490
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -1.82108 0.23306 -7.814 1.4e-12 ***
## waiting 0.07478 0.00323 23.148 < 2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.5183 on 135 degrees of freedom
## Multiple R-squared: 0.7988, Adjusted R-squared: 0.7973
## F-statistic: 535.8 on 1 and 135 DF, p-value: < 2.2e-1649.3 Predicting a New Value
So according to our model, what’s the predicted value of the eruption duration eruption given a waiting time of \(80\)?
Using the linear model: \[\hat{ED} = \hat{\beta_0} + \hat{\beta_1} WT\]
# with an example waiting time of 80, what is the expected value for eruption duration?
coef(lm1)[1] + coef(lm1)[2]*80## (Intercept)
## 4.161219## 1
## 4.16121949.4 Plot Predictions: Training vs Test Set
par(mfrow = c(1,2))
plot(trainFaith$waiting, trainFaith$eruptions, col = "red", xlab = "Waiting", ylab = "Eruption Duration", main = "Training Set")
lines(trainFaith$waiting, predict(lm1), lwd = 3)
plot(testFaith$waiting, testFaith$eruptions, col = "blue", xlab = "Waiting", ylab = "Eruption Duration", main = "Predictions on the Test Set")
lines(testFaith$waiting, predict(lm1, newdata = testFaith), lwd = 3)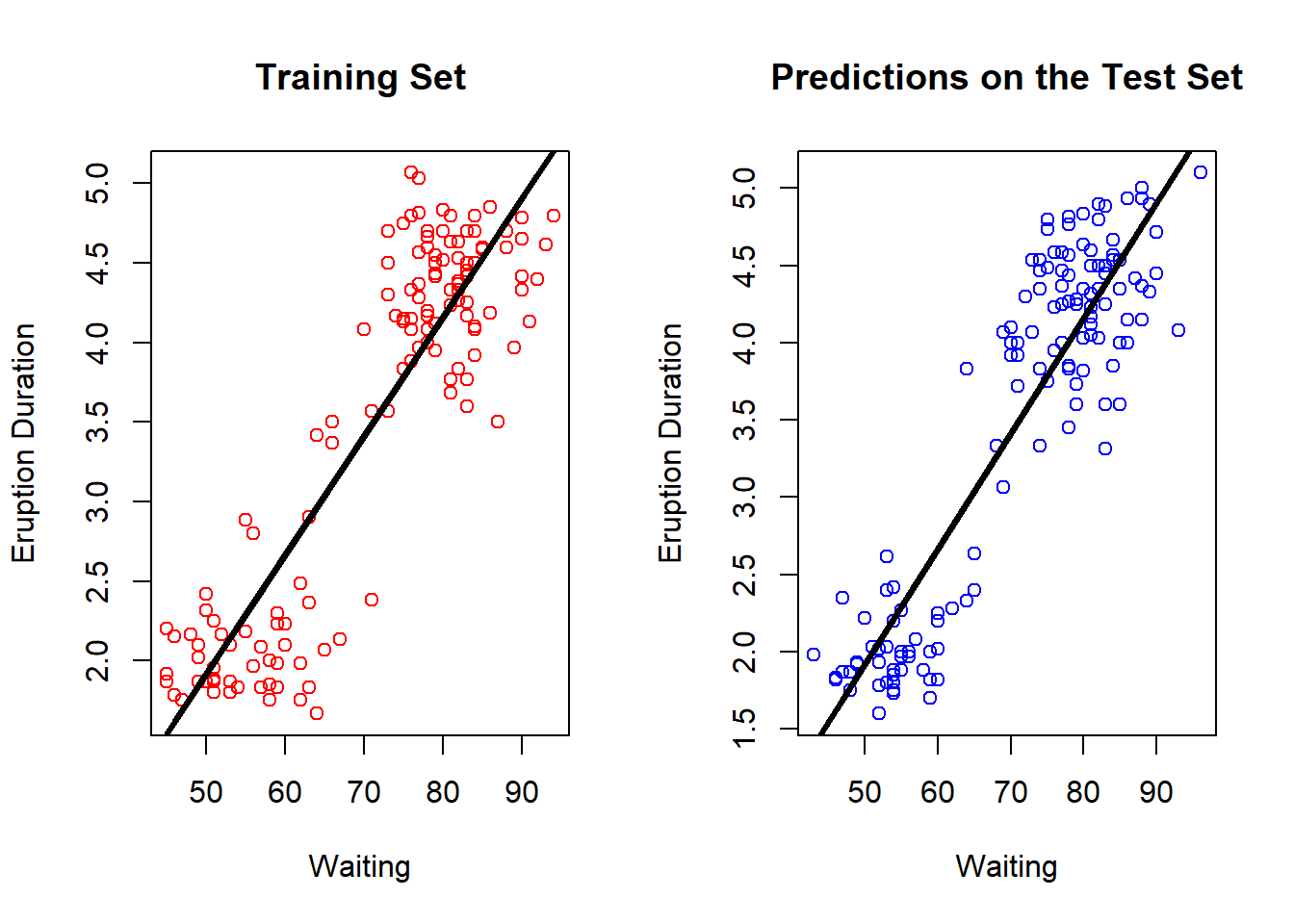
49.5 Get Training and Test set Errors
The measure of error for the linear model here will be \(RMSE\). This is the total difference between the fitted points and the actual points.
The \(RMSE\) for the test set is the exact same bench
## [1] 6.021535# Calculate the RMSE on the test set
sqrt(sum((predict(lm1, newdata = testFaith)-testFaith$eruptions)^2))## [1] 5.50939149.6 Prediction Intervals
Prediction intervals can be useful for looking at possible ranges that the values are likely to take. However, confidence intervals in general are probably more useful, this looks more or less intuitive going by the variation around the regression line.
Below we’ll use the regular plot(), matlines() and prediciton() functions, however this can also be done in the caret package.
# Calculate the prediction intervals! Using the linear model that we built on the training data
pred1 <- predict(lm1, newdata = testFaith, interval = "prediction")
ord <- order(testFaith$waiting)
plot(testFaith$waiting, testFaith$eruptions, pch = 8, col = "red")
matlines(testFaith$waiting[ord], pred1[ord, ], type = "l", col = c("blue","orange","orange"), lty = c(2,1,2), lwd = 3)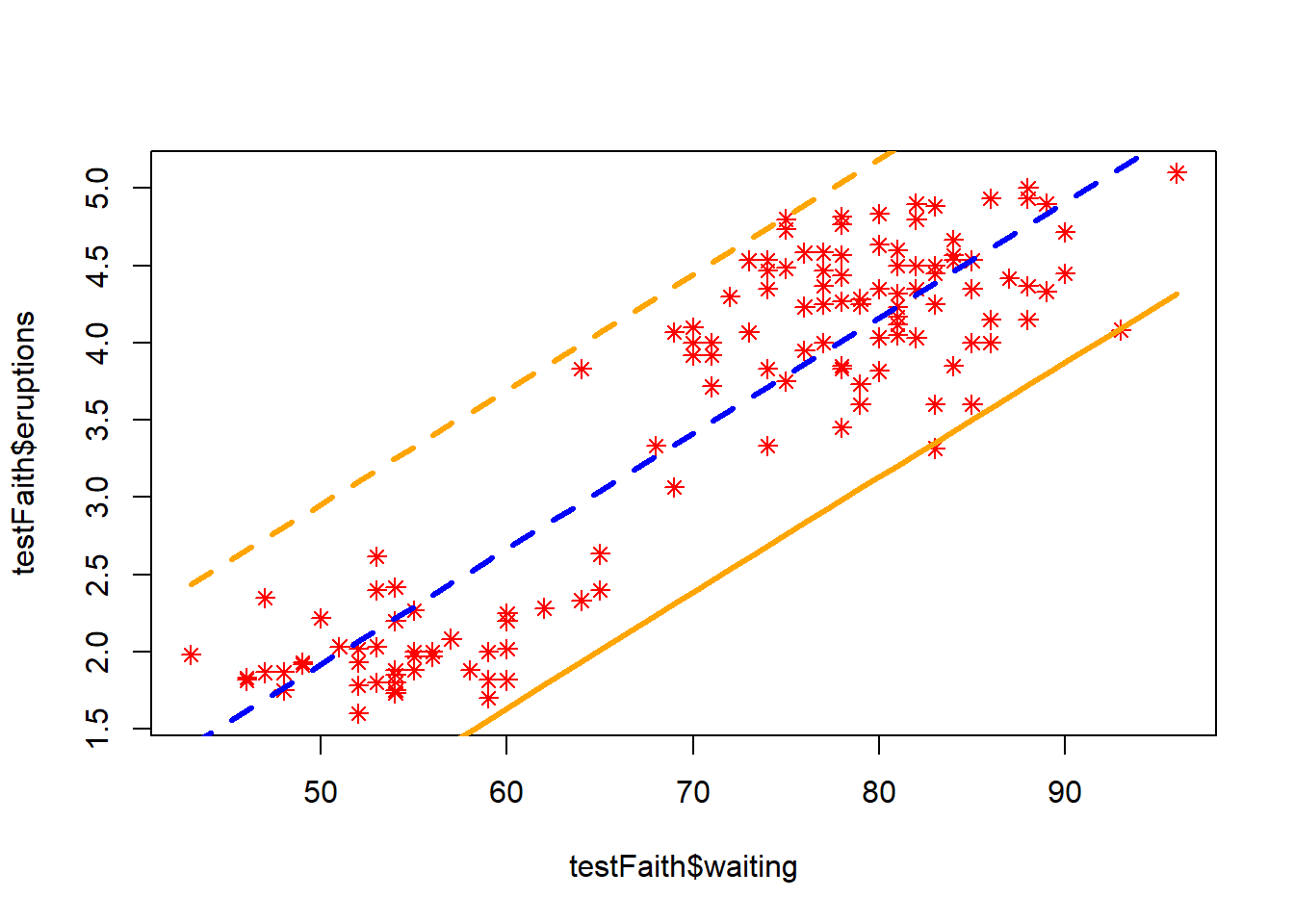
This process can also be done in the caret package.
# Create the model using the train function
modFit <- train(eruptions ~ waiting, data = trainFaith, method = "lm")
# The finalModel variable in the modFit object contains detailed information about the model created above, including the regression coefficients
summary(modFit$finalModel)##
## Call:
## lm(formula = .outcome ~ ., data = dat)
##
## Residuals:
## Min 1Q Median 3Q Max
## -1.29776 -0.37733 0.03967 0.35578 1.20490
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -1.82108 0.23306 -7.814 1.4e-12 ***
## waiting 0.07478 0.00323 23.148 < 2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.5183 on 135 degrees of freedom
## Multiple R-squared: 0.7988, Adjusted R-squared: 0.7973
## F-statistic: 535.8 on 1 and 135 DF, p-value: < 2.2e-16