54 Boosting
54.1 Basic Idea
- Take lots of potentially weak predictors.
- Weigh them and add them up.
- Get a stronger predictor.
- Start with a set of classifiers \(h_1, ..., h_k\)
- Examples: All possible trees, all possible regression models, all possible cut-offs.
- Create a classifier that combines classification functions
\[f(x) = sgn(\sum_{t=1}^T \alpha_t h_t(x))\]
- Goal is to minimise error (on training set)
- Iterative, select one \(h\) at each step
- Calculate weights based on errors
- Upweigh missed classifications and select \(h\)
- View Adaboost
54.2 Boosting in R
- Boosting can be done with any subset of classifiers
- One large subclass is gradient boosting
- R has multiple boosting libraries. Differences include the choice of basic classification functions and combination rules.
- gbm - boosting with trees
- mboost - model based boosting
- ada - statistical boosting based on additive logistic regression
- gamBoost for boosting generalised additive models
- Most of these are available in the caret package
54.3 Wage Example
library(gbm)
# Load Data
data(Wage)
# Subset and build training and testing sets
Wage <- subset(Wage, select = -c(logwage))
inTrain <- createDataPartition(y = Wage$wage ,
p = 0.7, list = FALSE)
training <- Wage[inTrain, ]
testing <- Wage[-inTrain, ]
# Model wage as a combination of all remaining variables
modFit <- train(wage ~ ., method = "gbm",
data = training, verbose = FALSE)
print(modFit)## Stochastic Gradient Boosting
##
## 2102 samples
## 9 predictor
##
## No pre-processing
## Resampling: Bootstrapped (25 reps)
## Summary of sample sizes: 2102, 2102, 2102, 2102, 2102, 2102, ...
## Resampling results across tuning parameters:
##
## interaction.depth n.trees RMSE Rsquared MAE
## 1 50 34.56805 0.3186570 23.32001
## 1 100 34.03865 0.3286803 22.96212
## 1 150 33.98890 0.3297850 22.98726
## 2 50 33.98977 0.3315167 22.92748
## 2 100 33.99139 0.3304338 23.01636
## 2 150 34.09998 0.3274460 23.14241
## 3 50 33.96126 0.3314366 22.95055
## 3 100 34.15682 0.3258006 23.19512
## 3 150 34.36910 0.3199571 23.40490
##
## Tuning parameter 'shrinkage' was held constant at a value of 0.1
##
## Tuning parameter 'n.minobsinnode' was held constant at a value of 10
## RMSE was used to select the optimal model using the smallest value.
## The final values used for the model were n.trees = 50, interaction.depth =
## 3, shrinkage = 0.1 and n.minobsinnode = 10.# View the results
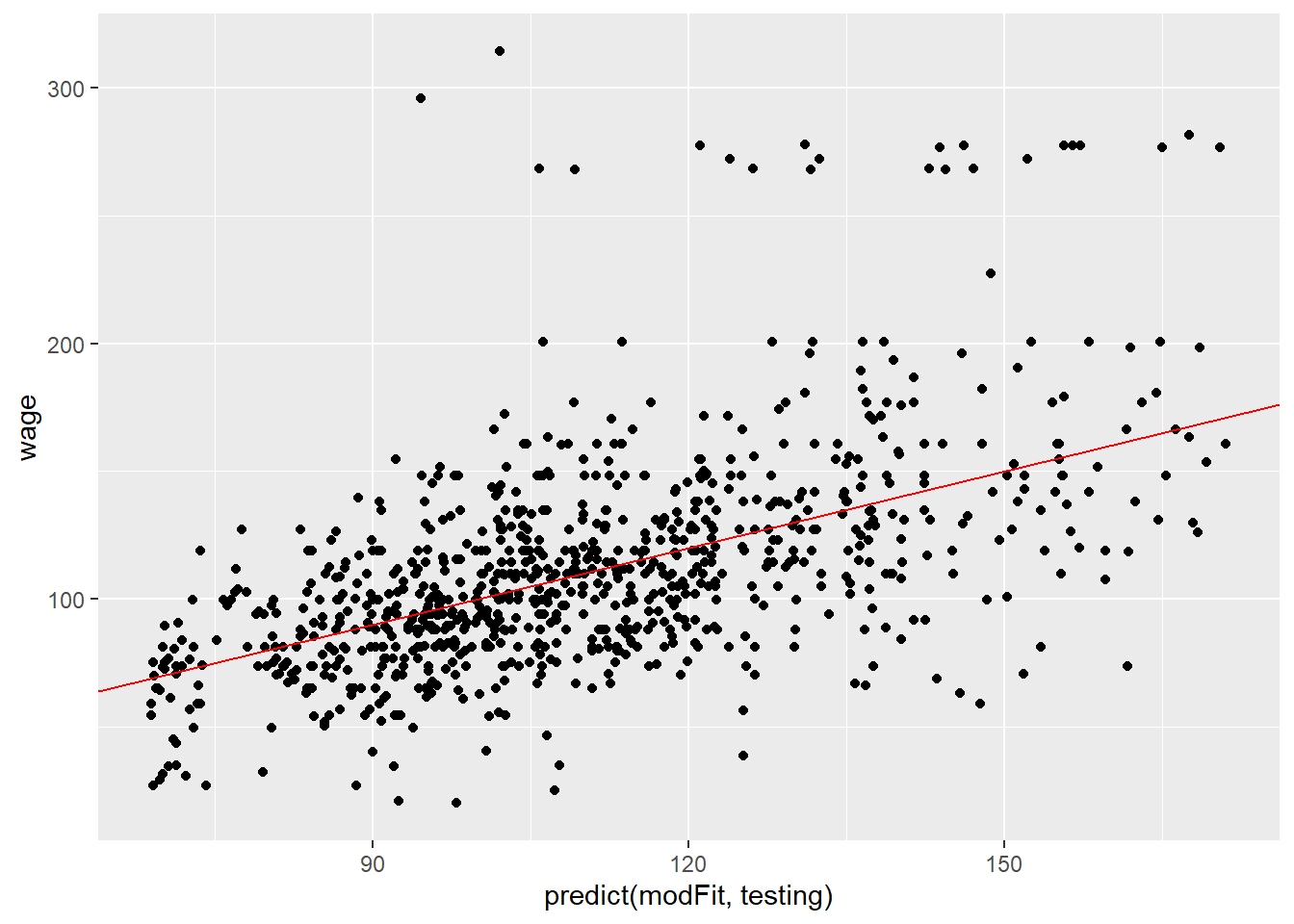
qplot(predict(modFit, testing), wage, data = testing) +
geom_abline(slope = 1, col = "red")